Write your own workflows - EoS example
Contents
6. Write your own workflows - EoS example#
Learning Objectives
In this section we will look at how you can write your own workflows.
We will also introduce the concepts of calcfunctions, workfunctions, and workchains, and how to use them to write a simple workflow for computing an Equation of State (EoS).
Lets look at a representative example of a workflow; a simple calculation of the Equation of State (EoS) of a crystal structure.
An equation of state consists of calculating the total energy (E) as a function of the unit cell volume (V). For this we need to relax the structure at a number of different volumes, and then analyse the results.
Show cell content
from local_module import load_temp_profile
data = load_temp_profile(name="workflow", add_computer=True, add_pw_code=True, add_sssp=True, add_structure_si=True)
data
AiiDALoaded(profile=Profile<uuid='6238b3e2b84e4fb1ba26912a656ca425' name='workflow'>, computer=<Computer: local_direct (localhost), pk: 1>, code=<Code: Remote code 'pw.x' on local_direct, pk: 1, uuid: ec4b5510-29d4-484f-ac04-b55d0bde8df0>, pseudos=SsspFamily<1>, structure=<StructureData: uuid: 7c33f04c-63d7-44bc-ac86-221486e25cd8 (pk: 87)>, cpu_count=1, workdir=PosixPath('/home/docs/checkouts/readthedocs.org/user_builds/aiida-qe-demo/checkouts/latest/tutorial/local_module/_aiida_workdir/workflow'), pwx_path=PosixPath('/home/docs/checkouts/readthedocs.org/user_builds/aiida-qe-demo/conda/latest/bin/pw.x'))
6.1. Parametrizing the input structure volume#
In the previous section on Generating and managing inputs we saw how AiiDA can interface with ASE. We can use this to write a simple function for generating a set of input structures.
from aiida import engine, orm
@engine.calcfunction
def rescale(structure: orm.StructureData, scale: orm.Float) -> orm.StructureData:
"""Rescale a structure's unit cell."""
from aiida.orm import StructureData
ase_structure = structure.get_ase()
scale_value = scale.value
new_cell = ase_structure.get_cell() * scale_value
ase_structure.set_cell(new_cell, scale_atoms=True)
return StructureData(ase=ase_structure)
Note the @engine.calcfunction function decorator.
This is a special decorator that tells AiiDA that this function should be treated as a calculation node in the provenance graph.
Now when we run this function, we will store the output structure in the profile, but also their link to the original input structure.
for factor in (0.99, 1.0, 1.01):
rescale(data.structure, orm.Float(factor))
from aiida.tools.visualization import Graph
graph = Graph(graph_attr={"rankdir": "LR", "size": "8!,8!"})
graph.recurse_descendants(
data.structure, depth=2, annotate_links="both", include_process_inputs=True
)
graph.graphviz
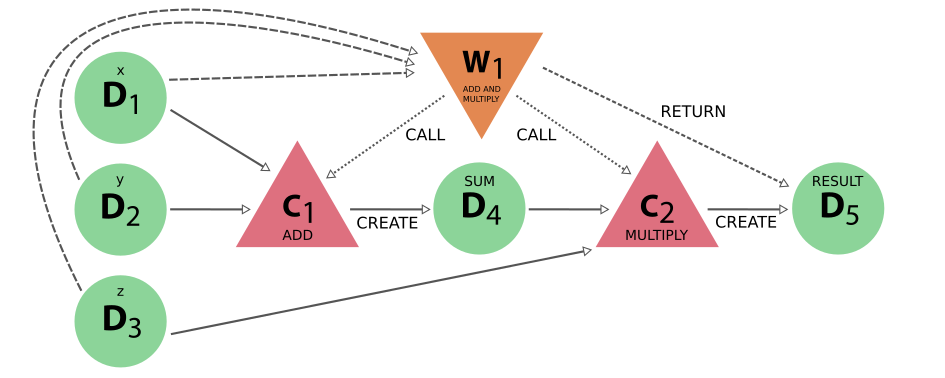
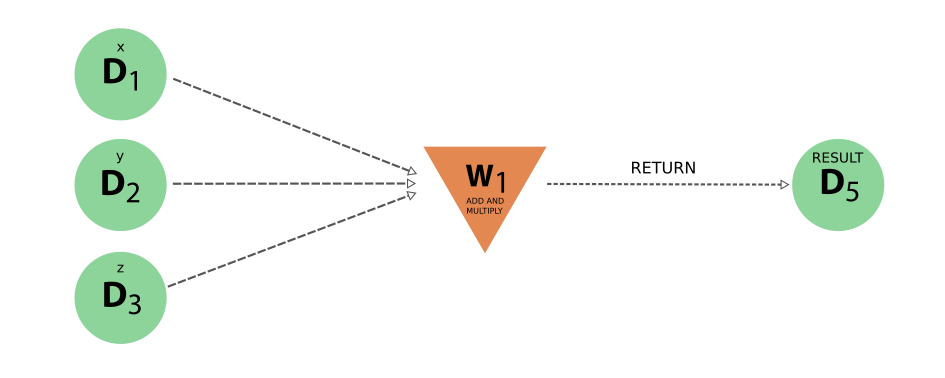
6.2. What’s the difference between a workflow and a calculation?#
In the calcfunction example above, we saw that we can use the @engine.calcfunction decorator to tell AiiDA that a function should be treated as a calculation node in the provenance graph.
A calculation is process which takes some data inputs, and produces some data outputs, i.e. it is a creator of data and can only have one step.
By contrast, a workflow is a process which does not itself create data, but rather calls child calculations to create the data, passing the intermediate result between them.
In this way, we can separate the logical flow of the computations from the physical flow of the data.
6.3. Writing an EoS workfunction#
First lets define some “helper” functions for setting up the SCF calculations, for each structure, and for bundling the results into a single dictionary.
Show cell source
from aiida import engine, orm
def generate_scf_input_params(
structure: orm.StructureData, code: orm.Code, pseudo_family
) -> engine.ProcessBuilder:
"""Construct a builder for the `PwCalculation` class and populate its inputs.
"""
parameters = {
"CONTROL": {
"calculation": "scf",
"tstress": True, # Important that this stays to get stress
"tprnfor": True,
},
"SYSTEM": {
"ecutwfc": 30.0,
"ecutrho": 200.0,
},
"ELECTRONS": {
"conv_thr": 1.0e-6,
},
}
kpoints = orm.KpointsData()
kpoints.set_kpoints_mesh([2, 2, 2])
builder = code.get_builder()
builder.code = code
builder.structure = structure
builder.kpoints = kpoints
builder.parameters = orm.Dict(dict=parameters)
builder.pseudos = pseudo_family.get_pseudos(structure=structure)
builder.metadata.options.resources = {"num_machines": 1}
builder.metadata.options.max_wallclock_seconds = 30 * 60
return builder
@engine.calcfunction
def create_eos_dictionary(**kwargs: orm.Dict) -> orm.Dict:
"""Create a single `Dict` node from the `Dict` output parameters of completed `PwCalculations`.
The dictionary will contain a list of tuples, where each tuple contains the volume, total energy and its units
of the results of a calculation.
"""
eos = [
(result.dict.volume, result.dict.energy, result.dict.energy_units)
for label, result in kwargs.items()
]
return orm.Dict(dict={"eos": eos})
Now we can write a simple EoS workfunction, which internally calls calculations to create the data, and then bundles the results into a single dictionary.
Similarly to the calcfunction example above, we use the @engine.workfunction decorator to tell AiiDA that this function should be treated as a workflow and store links between the nodes.
from aiida import engine, orm
from aiida_quantumespresso.calculations.pw import PwCalculation
@engine.workfunction
def run_eos_wf(
code: orm.Code, pseudo_family_label: orm.Str, structure: orm.StructureData
) -> orm.Dict:
"""Run an equation of state of a bulk crystal structure for the given element."""
# This will print the pk of the work function
print("Running run_eos_wf<{}>".format(engine.Process.current().pid))
scale_factors = (0.96, 0.98, 1.0, 1.02, 1.04)
labels = ["c1", "c2", "c3", "c4", "c5"]
pseudo_family = orm.load_group(pseudo_family_label.value)
calculations = {}
# Loop over the label and scale_factor pairs
for label, factor in list(zip(labels, scale_factors)):
# Generated the scaled structure from the initial structure
rescaled_structure = rescale(structure, orm.Float(factor))
# Generate the inputs for the `PwCalculation`
inputs = generate_scf_input_params(rescaled_structure, code, pseudo_family)
# Launch a `PwCalculation` for each scaled structure
print(
"Running a scf for {} with scale factor {}".format(
structure.get_formula(), factor
)
)
calculations[label] = engine.run(PwCalculation, **inputs)
inputs = {
label: result["output_parameters"] for label, result in calculations.items()
}
eos = create_eos_dictionary(**inputs)
return eos
We run the workfunction simply by calling it, and after we can see the process which has run in the CLI:
eos_wf_result = run_eos_wf(data.code, orm.Str(data.pseudos.label), data.structure)
eos_wf_result.get_dict()
Running run_eos_wf<98>
Running a scf for Si2 with scale factor 0.96
Running a scf for Si2 with scale factor 0.98
Running a scf for Si2 with scale factor 1.0
Running a scf for Si2 with scale factor 1.02
Running a scf for Si2 with scale factor 1.04
{'eos': [[34.007866582747, -307.37870009109, 'eV'],
[36.177946828062, -307.79009811293, 'eV'],
[38.438434271807, -308.06815096793, 'eV'],
[40.79117395684, -308.23441211264, 'eV'],
[43.238010927712, -308.3061811841, 'eV']]}
wf_node = eos_wf_result.base.links.get_incoming(orm.WorkFunctionNode).one().node
%verdi process status {wf_node.pk}
run_eos_wf<98> Finished [0]
├── rescale<100> Finished [0]
├── PwCalculation<104> Finished [0]
├── rescale<111> Finished [0]
├── PwCalculation<115> Finished [0]
├── rescale<122> Finished [0]
├── PwCalculation<126> Finished [0]
├── rescale<133> Finished [0]
├── PwCalculation<137> Finished [0]
├── rescale<144> Finished [0]
├── PwCalculation<148> Finished [0]
└── create_eos_dictionary<154> Finished [0]
From our results, we can visualise both the “logical”, “data” and “full” provenance of our workflow.
from aiida.tools.visualization import Graph
graph = Graph(graph_attr={"rankdir": "LR", "size": "8!,8!"})
graph.recurse_ancestors(
eos_wf_result, annotate_links="both", link_types=("input_work", "return")
)
graph.graphviz
graph = Graph(graph_attr={"rankdir": "LR", "size": "8!,8!"})
graph.recurse_ancestors(
eos_wf_result, annotate_links="both", link_types=("input_calc", "create")
)
graph.graphviz
Show cell output
graph = Graph(graph_attr={"rankdir": "LR", "size": "8!,8!"})
graph.recurse_ancestors(
eos_wf_result, annotate_links="both", include_process_outputs=True
)
graph.graphviz
Show cell output
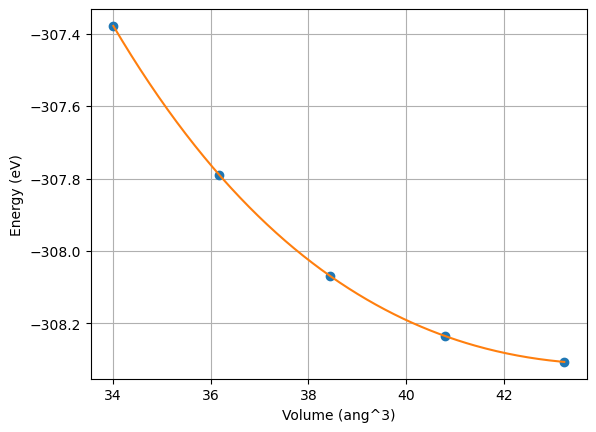
Then naturally we can also run post-analysis on the results, such as plotting the EoS.
Show cell source
from typing import List, Tuple
from matplotlib import pyplot as plt
import numpy as np
from scipy.optimize import curve_fit
def birch_murnaghan(V, E0, V0, B0, B01):
"""Compute energy by Birch Murnaghan formula."""
r = (V0 / V) ** (2.0 / 3.0)
return E0 + 9.0 / 16.0 * B0 * V0 * (r - 1.0) ** 2 * (2.0 + (B01 - 4.0) * (r - 1.0))
def fit_birch_murnaghan_params(volumes_, energies_):
"""Fit Birch Murnaghan parameters."""
volumes = np.array(volumes_)
energies = np.array(energies_)
params, covariance = curve_fit(
birch_murnaghan,
xdata=volumes,
ydata=energies,
p0=(
energies.min(), # E0
volumes.mean(), # V0
0.1, # B0
3.0, # B01
),
sigma=None,
)
return params, covariance
def plot_eos(eos: List[Tuple[float, float, str]]):
"""
Plots equation of state taking as input the pk of the ProcessCalculation
printed at the beginning of the execution of run_eos_wf
"""
data = np.array([(V, E) for V, E, _ in eos])
params, _covariance = fit_birch_murnaghan_params(data[:, 0], data[:, 1])
vmin = data[:, 0].min()
vmax = data[:, 0].max()
vrange = np.linspace(vmin, vmax, 300)
fig, ax = plt.subplots()
ax.plot(data[:, 0], data[:, 1], "o")
ax.plot(vrange, birch_murnaghan(vrange, *params))
ax.grid(True)
ax.set_xlabel("Volume (ang^3)")
ax.set_ylabel("Energy ({})".format(eos[0][2]))
return fig, ax
fig, ax = plot_eos(eos_wf_result.get_dict()["eos"])
6.4. Writing an EoS workchain#
In the run_eos_wf example above, we encapsulate all the logic of the workflow in a single function.
This has the advantage of being simple to write, but also limitations:
The structure relaxations are run synchronously, i.e. one after the other. This means that the workflow will not scale well to large numbers of structure volumes.
The workflow is not restartable, i.e. if it is interrupted, it will have to start from scratch.
To overcome these limitations, we can instead write a WorkChain class, which allows us to:
Run the calculations asynchronously, i.e. in parallel.
Write the workflow logic in a modular way, i.e. as a series of steps.
Write the workflow logic in a way that allows it to be restarted from any step. Each step stores a checkpoint in the profile storage, which can be used to restart the workflow from that point, if for example you shut down the machine on which AiiDA is running.
from aiida import engine, orm
from aiida_quantumespresso.calculations.pw import PwCalculation
scale_facs = (0.96, 0.98, 1.0, 1.02, 1.04)
labels = ["c1", "c2", "c3", "c4", "c5"]
class EquationOfState(engine.WorkChain):
"""WorkChain to compute Equation of State using Quantum ESPRESSO."""
@classmethod
def define(cls, spec: engine.ProcessSpec):
"""Specify inputs and outputs."""
super().define(spec)
spec.input("code", valid_type=orm.Code)
spec.input("pseudo_family_label", valid_type=orm.Str)
spec.input("structure", valid_type=orm.StructureData)
spec.outline(
cls.run_eos,
cls.results,
)
spec.output("eos", valid_type=orm.Dict)
def run_eos(self):
"""Run calculations for equation of state."""
# Create basic structure and attach it as an output
structure = self.inputs.structure
pseudo_family = orm.load_group(self.inputs.pseudo_family_label.value)
for label, factor in zip(labels, scale_facs):
rescaled_structure = rescale(structure, orm.Float(factor))
inputs = generate_scf_input_params(
rescaled_structure, self.inputs.code, pseudo_family
)
self.report(
"Running an SCF calculation for {} with scale factor {}".format(
structure.get_formula(), factor
)
)
# Ask the workflow to continue when the results are ready,
# and store them in the context
calc_future = self.submit(PwCalculation, **inputs)
self.to_context(**{label: calc_future})
def results(self):
"""Process results."""
inputs= {}
for label in labels:
calcnode: orm.CalcJobNode = self.ctx[label]
inputs[label] = calcnode.base.links.get_outgoing().get_node_by_label("output_parameters")
eos = create_eos_dictionary(**inputs)
# Attach Equation of State results as output node to be able to plot the EOS later
self.out("eos", eos)
Of note in the workchain above are the following elements:
The
defineclass method, which specifies the inputs and outputs of the workchain, and the steps of the workflow.The
self.reportmethod, which allows us to log messages about the process.The
self.to_contextmethod andself.ctxattribute, which allows us to store data between steps of the workflow, and access it from other steps.The context is persisted in the profile storage, and so can be used to restart the workflow from any step.
The
self.submitmethod, which allows us to gather calculations for execution, and return aFutureobject which can be used to access the results of the calculation.Only at the end of the workchain step will the calculations be submitted, then the workchain will wait for them to finish before continuing to the next step.
When the workchain is submitted to the daemon, this will handle distributing the calculations to the available workers.
eos_wc_result = engine.run_get_node(
EquationOfState,
code=data.code,
pseudo_family_label=orm.Str(data.pseudos.label),
structure=data.structure,
)
eos_wc_result.node
Report: [157|EquationOfState|run_eos]: Running an SCF calculation for Si2 with scale factor 0.96
Report: [157|EquationOfState|run_eos]: Running an SCF calculation for Si2 with scale factor 0.98
Report: [157|EquationOfState|run_eos]: Running an SCF calculation for Si2 with scale factor 1.0
Report: [157|EquationOfState|run_eos]: Running an SCF calculation for Si2 with scale factor 1.02
Report: [157|EquationOfState|run_eos]: Running an SCF calculation for Si2 with scale factor 1.04
<WorkChainNode: uuid: 31319886-2475-42da-949a-ab4a54569f80 (pk: 157) (__main__.EquationOfState)>
%verdi process status {eos_wc_result.node.pk}
EquationOfState<157> Finished [0] [1:results]
├── rescale<159> Finished [0]
├── PwCalculation<163> Finished [0]
├── rescale<165> Finished [0]
├── PwCalculation<169> Finished [0]
├── rescale<171> Finished [0]
├── PwCalculation<175> Finished [0]
├── rescale<177> Finished [0]
├── PwCalculation<181> Finished [0]
├── rescale<183> Finished [0]
├── PwCalculation<187> Finished [0]
└── create_eos_dictionary<213> Finished [0]
%verdi process show {eos_wc_result.node.pk}
Show cell output
Property Value
----------- ------------------------------------
type EquationOfState
state Finished [0]
pk 157
uuid 31319886-2475-42da-949a-ab4a54569f80
label
description
ctime 2022-10-04 08:16:27.847375+00:00
mtime 2022-10-04 08:16:43.629186+00:00
computer [1] local_direct
Inputs PK Type
------------------- ---- -------------
code 1 Code
pseudo_family_label 156 Str
structure 87 StructureData
Outputs PK Type
--------- ---- ------
eos 214 Dict
Called PK Type
-------- ---- ---------------------
CALL 159 rescale
CALL 163 PwCalculation
CALL 165 rescale
CALL 169 PwCalculation
CALL 171 rescale
CALL 175 PwCalculation
CALL 177 rescale
CALL 181 PwCalculation
CALL 183 rescale
CALL 187 PwCalculation
CALL 213 create_eos_dictionary
Finally, we recreate out EoS plot from the results of the workchain 🎉
fig, ax = plot_eos(eos_wc_result.result["eos"].get_dict()["eos"])

6.5. Utilising WorkChains in the daemon#
In the previous example, we ran the workchain in the foreground, i.e. we waited for it to finish before continuing to the next step.
In practice, we will want to run the workchain in the background, i.e. submit it to the daemon, and then continue with other work.
For the daemon to be able to find the workchain, we need to register it with AiiDA.
This entails either adding the python file containing the workchain to the PYTHONPATH environment variable,
or better, creating a Python package and installing it in the environment.
See the aiida-plugin-cutter template, and aiida-diff exemplar plugin for how to do this.
Important
After any changes to the source code of the workchain, you will need to restart the daemon for it to load the new code.
$ verdi daemon restart --reset